Publications
Group highlights
(For a full list see below or go to Google Scholar.)
M. Benton, M. Furr, V. Govind Kumar, A. Polasa, F. Gao, C-D Heyes, T-K Suresh Kumar, M. Moradi

J.Chem. Inf. Model, vol. 63, pp. 4125–4137, 2023
A. Polasa, I. Mosleh, J. Losey, A. Abbaspourrad, R. Beitle and M. Moradi
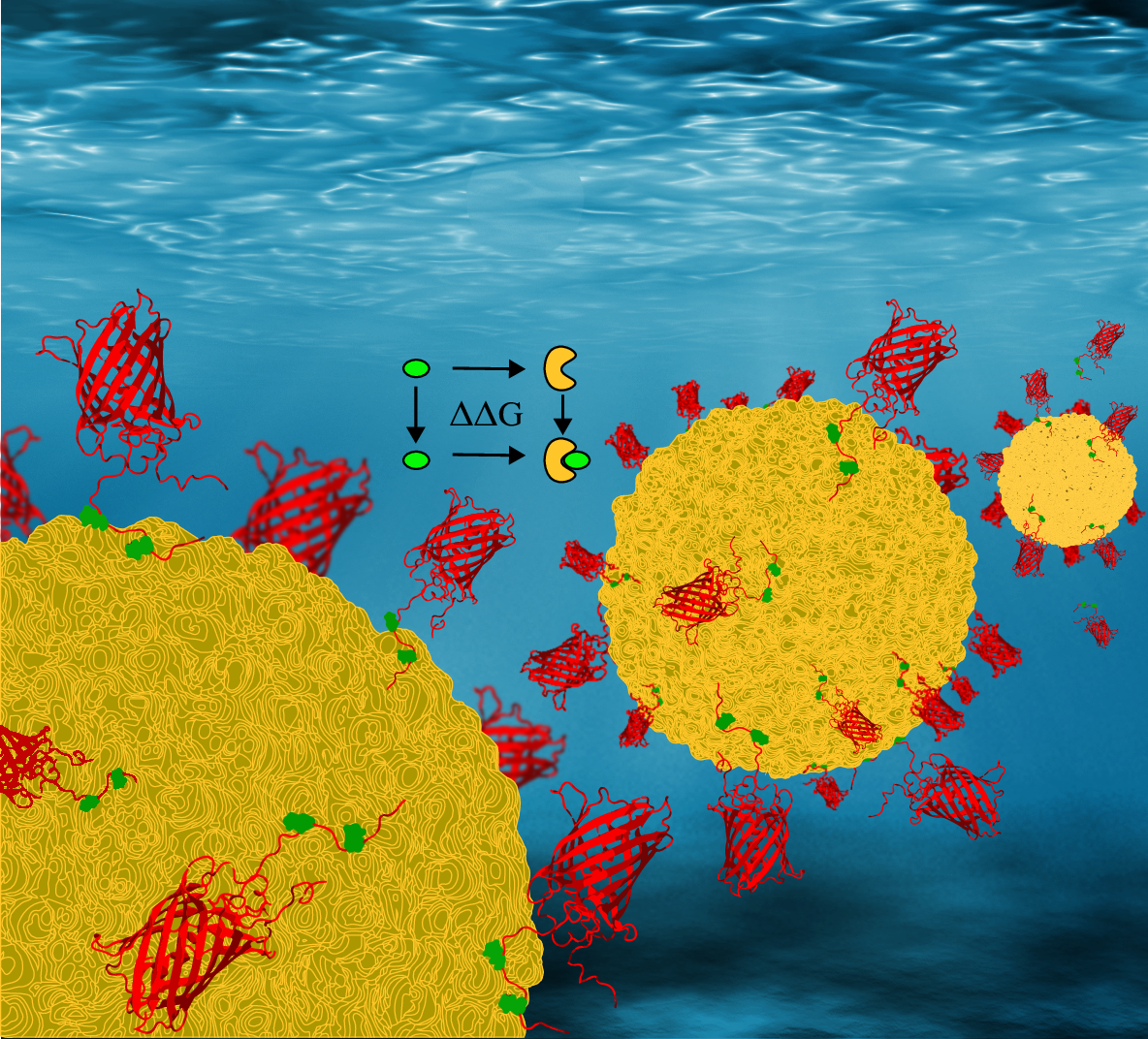
Nanoscale Adv, vol. 4, pp. 3161–3171, 2022
S. Sauve, J.Williamson, A. Polasa and M. Moradi
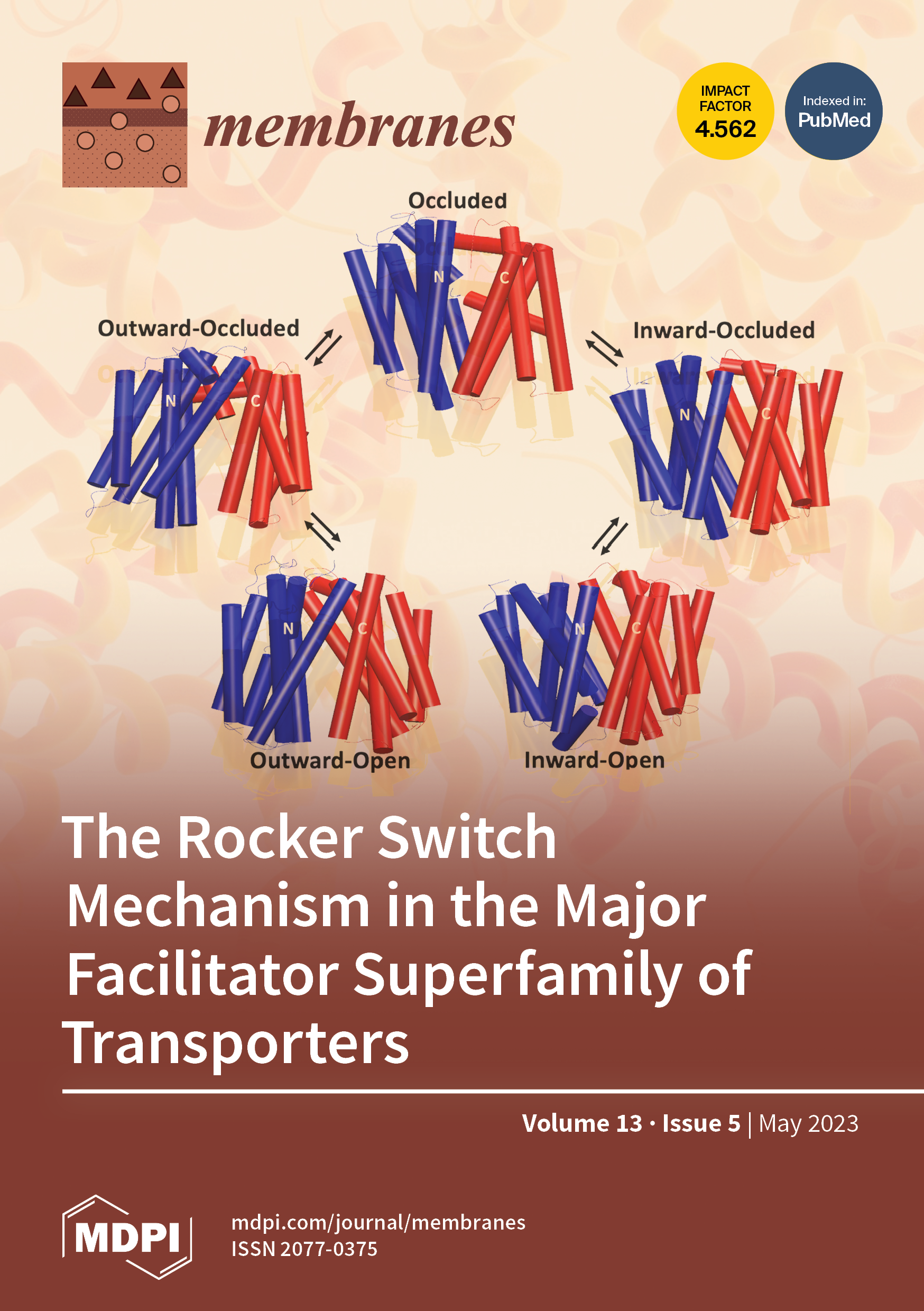
Membranes, vol. 13, pp. 462-479, 2023
V. Govind. Kumar, D. S. Ogden, U. H. Isu, A. Polasa, J. Losey and M. Moradi
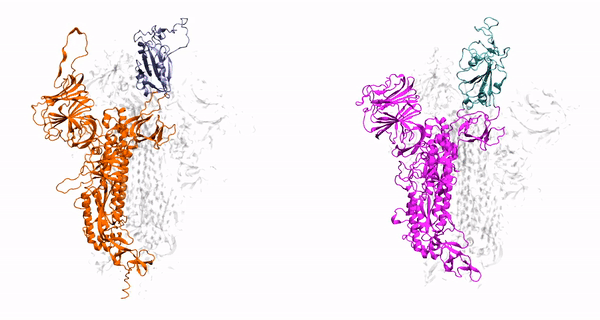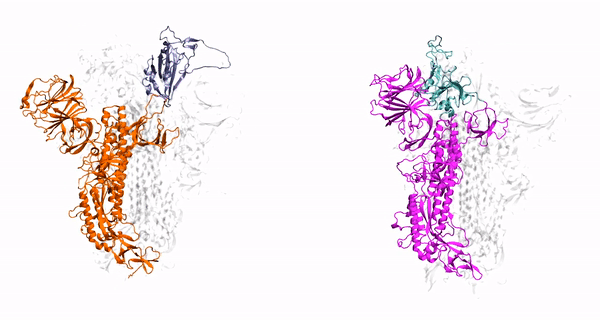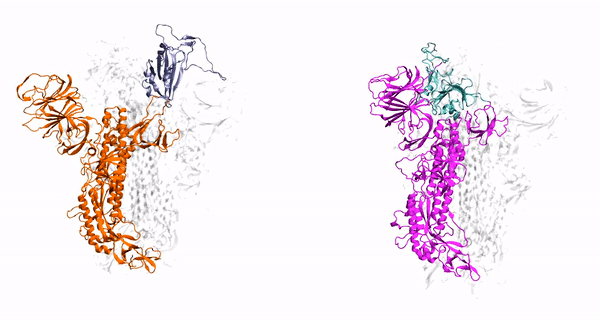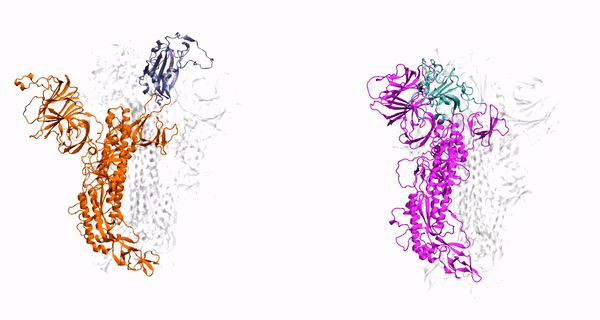
J. Biol. Chem, vol. 298, pp. 1-12, 2022
Full List
2026
- A Fakharzadeh, C Goolsby, E Tajkhorshid, M Moradi, Thermodynamic and Kinetic Analysis of Molecular Conformational Dynamics in a Riemannian Framework The Journal of Physical Chemistry A, 2026
- J-L Losey, M Jauch,A Cortes-Cubero, H Wu, A Polasa, S Sauve, R Rivera, D-S Matteson, M Moradi, Simulating freely-diffusing single-molecule FRET data with consideration of protein conformational dynamics The Journal of Physical Chemistry B, 2026
- S Sauve, E Khodadadi, A Shubbar, E Khodadadi and M Moradi, Integrating Molecular Dynamics Simulations and Single-molecule FRET Spectroscopy: From Computational FRET Estimation to Experimental Data Interpretation The Journal of Physical Chemistry B, 130 (2), 651-667, 2026
Preprint
- R Dastvan, B Abd Emami, A Shubbar, H Woods, and M Moradi, Structural Dynamics of Sphingosine Kinase 1 Regulation and Inhibition Research Square
- E Khodadadi, E Khodadadi,P Chaturvedi and M Moradi, Molecular Insights into Cholesterol Concentration Effects on Planar and Curved Lipid Bilayers for Liposomal Drug Delivery bioRxiv
- E Khodadadi, S Badiee, E Khodadadi and M Moradi, Lipid-Mediated Modulation of mGluR2 Embedded in Micelle and Bilayer Environments: Insights from Molecular Dynamics bioRxiv
- S Badiee, J Hettige and M Moradi, Lipid-dependent conformational dynamics of bacterial ATP-binding cassette transporter Sav1866 bioRxiv
- U Isu, S Badiee, A Polasa, S H Tabari, M Derakhshani-Molayousefi and M Moradi, Cholesterol Dependence of the Conformational Changes in Metabotropic Glutamate Receptor 1 bioRxiv
- D Ogden and M Moradi, Atomic-level characterization of the conformational transition pathways in SARS-CoV-1 and SARS-CoV-2 spike proteins bioRxiv
- D Ogden, K Immadisetty, S Sauve, M Moradi, Conformational Transition Pathways in Major Facilitator Superfamily Transporters bioRxiv
2025
- E Khodadadi, E Khodadadi,P Chaturvedi and M Moradi, Molecular Dynamics Simulations of Liposomes: Structure, Dynamics, and Applications Membranes, 15 (9), 259, 2025
- A Fakharzadeh, M Moradi, C Sagui and C Roland, Comparative Study of the Bending Free Energies of C- and G‑Based DNA: A‑, B‑, and Z‑DNA and Associated Mismatched Trinucleotide Repeats, Journal of Chemical Information and Modeling, 65 (5672–5689), 2025
- S Thangapandian, A Fakharzadeh, M Moradi and E Tajkhorshid, Conformational free energy landscape of a glutamate transporter and microscopic details of its transport mechanism. Proceedings of the National Academy of Sciences, 122 (10), 2025
- E Khodadadi, E Khodadadi,P Chaturvedi and M Moradi, Comprehensive Insights into the Cholesterol-Mediated Modulation of Membrane Function through Molecular Dynamics Simulations Membranes, 15 (6), 173, 2025
2024
- M Furr, S A Badiee, S Basha, S Agrawal, Z Alraawi, S Heng, C L Stacy, Y Ahmed, M Moradi, T K S Kumar, R M Ceballos Structural stability comparisons between natural and engineered group II chaperonins: Are crenarchaeal heat shock proteins also pH shock resistant? Microorganisms, 12:2348, 2024.
- S Badiee, V Govind Kumar and M Moradi, Molecular dynamics investigation of the influenza hemagglutinin conformational changes in acidic pH The Journal of Physical Chemistry B, 128:11151, 2024.
- R Zhang , K Jagessar , M Brownd , A Polasa , R Stein , M Moradi ,E Karakas and H Mchaourab. Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling Nature Communications, 15:9055, 2024.
- A Polasa, S Badiee and M Moradi. Deciphering the Inter-domain Coupling in a Gram-negative Bacterial Membrane Insertase The Journal of Physical Chemistry B, 128:9734, 2024.
- U Isu, A Polasa and M Moradi. Differential Behavior of Conformational Dynamics in Active and Inactive States of Cannabinoid Receptor 1 The Journal of Physical Chemistry B, 128:8437, 2024.
- Y Ao, J R Grover, L Gifford, Y Han, G Zhong, R Katte, W Li, R Bhattacharjee, B Zhang, S Sauve, W Qin, D Ghimire, M A Haque, J Arthos, M Moradi, W Mothes, E A Lemke, P D Kwong, G B Melikyan, and M Lu. Bioorthogonal click labeling of an amber-free HIV-1 provirus for in-virus single molecule imaging Cell Chemical Biology, 31:487, 2024.
2023
- A Polasa and M Moradi, Towards A Purely Physics-based Computational Binding Affinity Estimation Nature Computational Science, 3:11, 2023.
- U Isu, S Badiee, E Khodadadi, M Moradi, Cholesterol in Class C GPCRs: Role, Relevance, and Localization Membranes, 13:301, 2023.
- S Sauve, J Williamson, A Polasa, M Moradi, Ins and Outs of Rocker Switch Mechanism in Major Facilitator Superfamilty of Transporters Membranes, 13:462, 2023.
- S Badiee, U Isu, E Khodadadi, M Moradi, The Alternating Access Mechanism in Mammalian Multidrug Resistance Transporters and Their Bacterial Homologs Membranes, 13:568, 2023.
- V Govind Kumar, A Polasa, S Agrawal, T K S Kumar, M Moradi, Binding Affinity Estimation from Restrained Umbrella Sampling Simulations Nature Computational Science, 3:59, 2023.
- M Benton, M Furr, V Govind Kumar, A Polasa, F Gao, C-D Heyes, T-K Suresh Kumar, M Moradi cpSRP43 is both highly flexible and stable: Structural insights using a combined experimental and computational approach Journal of Chemical Information and Modeling, 63 :4137, 2023.
- C Goolsby, J Losey, A Fakharzadeh, Y Xu, M Düker, M Getmansky Sherman, D Matteson, M Morad. Addressing the Embeddability Problem in Transition Rate Estimation The Journal of Physical Chemistry A, 127:5759, 2023.
2022
- A Polasa, J Hettige, K Immadisetty, M Moradi, An investigation of the YidC-mediated membrane insertion of Pf3 coat protein using molecular dynamics simulations Front. Mol. Biosci., 9 :15, 2022
- A Polasa, I Mosleh, J Losey, A Abbaspourrad, R Beitle, M Moradi, Developing a Rational Approach to Designing Recombinant Proteins for Peptide-Directed Nanoparticle Synthesis Nanoscale Adv., 4:3171, 2022
- K Immadisetty, A Polasa, R Shelton, and M Moradi, Elucidating the Molecular Basis of Spontaneous Activation in an Engineered Mechanosensitive Channel. Comput. Struct. Biotechnol. J., 20:2539, 2022.
- V Govind Kumar, D-S Ogden, U-H Isu, A Polasa, J-L Losey, and M Moradi, Prefusion Spike Protein Conformational Changes Are Slower in SARS-CoV-2 than in SARS-CoV-1. J. Biol. Chem, 298:101814, 2022.
- H Chen, D Ogden, S Pant, W Cai, E Tajkhorshid, M Moradi, B Roux, and C Chipot, A Companion Guide to the String Method with Swarms of Trajectories: Characterization, Performance, and Pitfalls J. Chem. Theory Comput., 18:1406–1422, 2022.
2021
- V Govind Kumar, S Agrawal, T-K Suresh Kumar, M Moradi, Mechanistic Picture for Monomeric Human Fibroblast Growth Factor 1 Stabilization by Heparin Binding. J. Phys. Chem. B, 125:12690-12697, 2021.
- D-R Baucom, M Furr, V Govind Kumar, P Okoto, J-L Losey, R-L Henry, M Moradi, T-K Suresh Kumar, C-D Heyes Transient local secondary structure in the intrinsically disordered C-term of the Albino3 insertase Biophys. J, 120:4992-5004, 2021.
- K Immadisetty and M Moradi Mechanistic Picture for Chemomechanical Coupling in a Bacterial Proton-Coupled Oligopeptide Transporter from Streptococcus Thermophilus J. Phys. Chem. B, 125:9738–9750, 2021.
- D Ogden, M Moradi, Molecular Dynamics–Based Thermodynamic and Kinetic Characterization of Membrane Protein Conformational Transitions Structure and Function of Membrane Proteins. Methods in Molecular Biology, 2302:309, 2021.
- S Agrawal, V Govind Kumar, R-K Gundampati, M Moradi & T-K Suresh Kumar, Characterization of the structural forces governing the reversibility of the thermal unfolding of the human acidic fbroblast growth factor Sci. Rep., 11:15579, 2021.
2019
- T Harkey, V Govind Kumar, J Hettige, S H Tabari, K Immadisetty, and M Moradi, The Role of a Crystallographically Unresolved Cytoplasmic Loop in Stabilizing the Bacterial Membrane Insertase YidC2. Sci. Rep., 9:14451, 2019.
- R Al Faouri, E Krueger, V Kumar, D Fologea, D Straub, H Alismail, Q Alfaori, A Kight, J Ray, R Henry, M Moradi, and G Salamo, An Effective Electric Dipole Model for Voltage-induced Gating Mechanism of Lysenin. Sci. Rep., 9:11440, 2019.
- K Immadisetty, J Hettige, and M Moradi, Lipid-Dependent Alternating Access Mechanism of a Bacterial Multidrug ABC Exporter. ACS Cent. Sci., 5:43-56, 2019.
2018
- S Burgin, J Oramous, M Kaminski, L Stocker, and M Moradi, High School Biology Students Use of VMD as an Authentic Tool for Learning about Modeling as a Professional Scientific Practice. Biochem. Mol. Biol. Educ, 46:230-236, 2018.
- M Kiaei, M Balasubramaniam, V Govind Kumar, R J Shmookler Reis, M Moradi, and K I Varughese, ALS-causing mutations in profilin-1 alter its conformational dynamics: A computational approach to explain propensity for aggregation. Sci. Rep., 2018 8:13102, 2018.
2017
- K Immadisetty, J Hettige, and M Moradi, What Can and Cannot Be Learned from Molecular Dynamics Simulations of Bacterial Proton-Coupled Oligopeptide Transporter GkPOT? J. Phys. Chem. B, 121:3644-3656, 2017.
- A Singharoy, C Chipot, M Moradi, and K Schulten, Chemomechanical coupling in hexameric protein–protein interfaces harness energy within V–type ATPases. J. Am. Chem. Soc., 139:293-310, 2017.
2016
- A Fakharzadeh and M Moradi, Effective Riemannian Diffusion Model for Conformational Dynamics of Biomolecular Systems. J. Phys. Chem. Lett., 7:4980-4987, 2016.
2015
- M Moradi, G Enkavi, and E Tajkhorshid, Atomic-level characterization of transport cycle thermodynamics in the glycerol-3-phosphate:phosphate antiporter. Nat. Commun., 6:8393, 2015.
- J Li, P-C Wen, M Moradi, and E Tajkhorshid, Computational Characterization of Structural Dynamics Underlying Function in Active Membrane Transporters. Curr. Opin. Struct. Biol., 31:96, 2015
- M Moradi, V Babin, C Roland, and C Sagui, The Adaptively Biased Molecular Dynamics method revisited: New capabilities and an application. J. Phys. Conf. Ser. 640:012020, 2015.
- M Moradi, C Sagui, and C Roland, Calculating transition and reaction rates with nonequilibrium work measurements. J. Phys. Conf. Ser. 640:012014, 2015.
2014
- M Moradi, C Sagui, and C Roland, Investigating rare events with nonequilibrium work measurements: I. nonequilibrium transition path probabilities. J. Chem. Phys., 140:034114, 2014.
- M Moradi, C Sagui, and C Roland, Investigating rare events with nonequilibrium work measurements: II. transition and reaction rates. J. Chem. Phys., 140:034115, 2014.
- R E Hulse, J Sachleben, P-C Wen, M Moradi, E Tajkhorshid, E Perozo, Conformational dynamics at the inner gate of KcsA during activation. Biochemistry (Rapid Report), 53:2557, 2014.
- M Moradi and E Tajkhorshid, Computational recipe for efficient description of large-scale conformational changes in biomolecular systems. J. Chem. Theory Comput., 10:2866, 2014.
- G Enkavi, J Li, P-C Wen, S Thangapandian, M Moradi, T Jiang, W Han, and E Tajkhorshid, A microscopic view of the mechanisms of active transport across the cellular membrane. Annual Reviews in Computational Chemistry, 10:77, 2014.
2013
- M Moradi and E Tajkhorshid,
Mechanistic picture for conformational transition of a membrane transporter at atomic resolution.
Proc. Natl. Aca. Sci. USA, 110:18916, 2013.



- M Moradi, and E Tajkhorshid, Driven metadynamics: Reconstructing equilibrium free energies from driven adaptive-bias simulations. J. Phys. Chem. Lett., 4:1882, 2013.
- M Moradi, V Babin, C Roland, and C Sagui, Reaction path ensemble of B-Z-DNA transition: A Comprehensive atomistic study. Nucleic Acids Res., 41:33, 2013.
- M Moradi, V Babin, C Sagui, and C Roland, Recipes for free energy calculations in biomolecular systems. In Biomolecular simulations: methods and protocols. Humana Press (Springer), Methods Mol. Biol., 924:313-37, 2013.
- M Moradi, V Babin, C Roland, and C Sagui, Free energies, structural characteristics, and transition mechanisms of proline-rich peptides. In Proline: Biosynthesis, Regulation and Health Benefits, Nova Publishers, ISBN: 978-1-62257-745-3, 2013.
- M Kalani, A Moradi, M Moradi, E Tajkhorshid, Characterizing a histidine switch controlling pH-dependent conformational changes of the influenza virus hemagglutinin. Biophys. J., 105:993, 2013.
2009-2012
- M Moradi, V Babin, C Roland, T Darden, and C Sagui, Conformations and free energy landscapes of polyproline peptides. Proc. Natl. Aca. Sci. USA, 106:20746, 2009.
- V Babin, V Karpusenka, M Moradi, C Roland, and C Sagui, Adaptively biased molecualr dynamics: An umbrella sampling method with a time-dependent potential. Int. J. Quant. Chem., 109:3666, 2009.
- M Moradi, V Babin, C Roland, and C Sagui, A classical molecular dynamics investigation of the free energy and structure of short polyproline conformers. J. Chem. Phys., 133:125104, 2010.
- M Moradi, J-G Lee, V Babin, C Roland, and C Sagui, Free energy and structure of polyproline peptides: an ab initio and classical molecular dynamics investigation. Int. J. Quant. Chem., 110:2865, 2010.
- M Moradi, V Babin, C Sagui, and C Roland, A statistical analysis of the PPII propensity of amino acid guests in proline-rich peptides. Biophys. J., 100:1083, 2011.
- M Moradi, V Babin, C Sagui, and C Roland, PPII propensity of multiple-guest amino acids in a proline-rich environment. J. Phys. Chem. B, 115:8645, 2011.
- M Moradi, C Sagui, and C Roland, Calculating relative transition rates with driven nonequilibrium simulations. Chem. Phys. Lett., 518:109, 2011.
- M Moradi, V Babin, C Roland, and C Sagui, Are long-range structural correlations behind the aggregation phenomena of polyglutamine diseases? PLoS Comput. Biol., 8:e1002501, 2012.