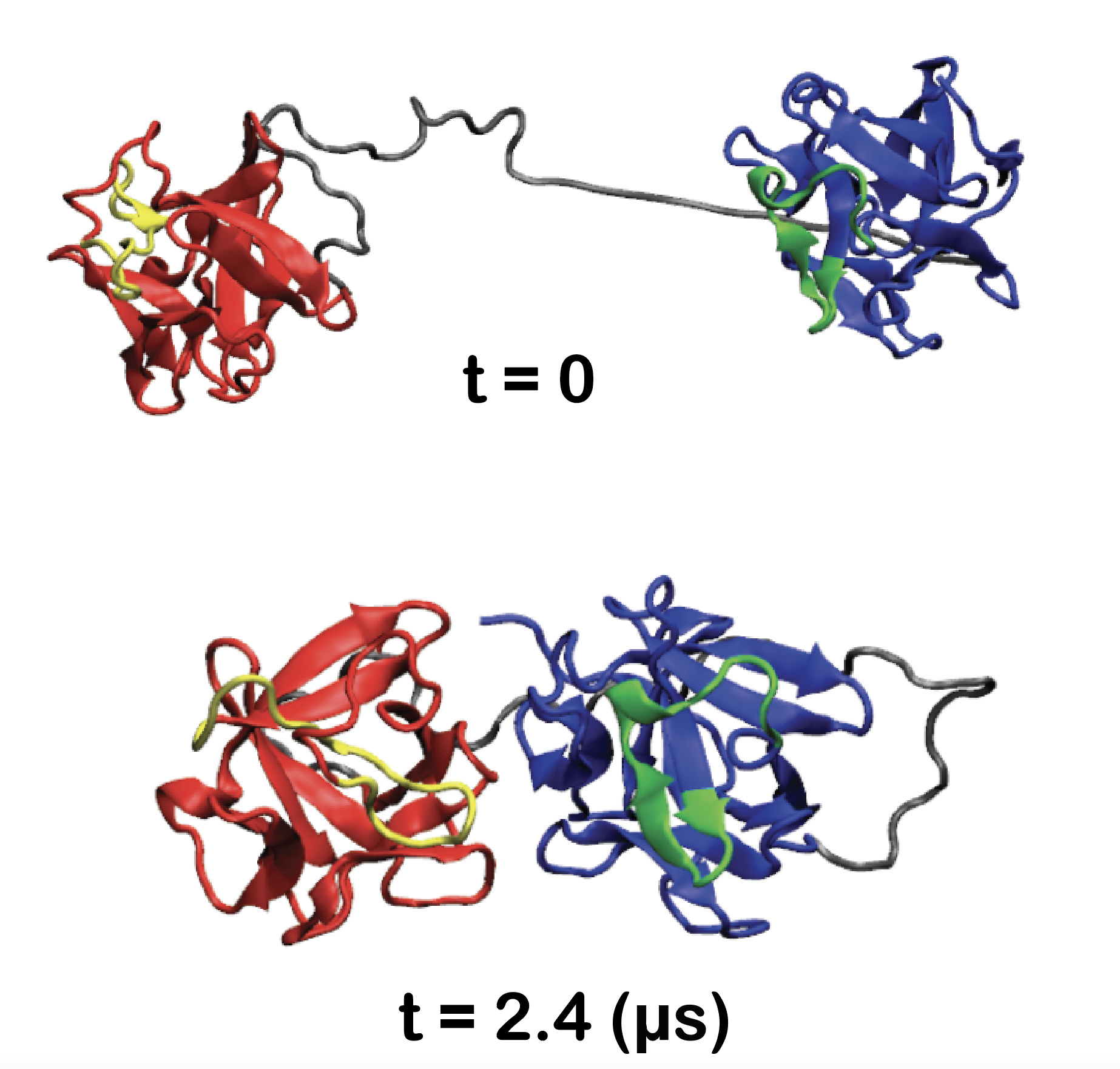
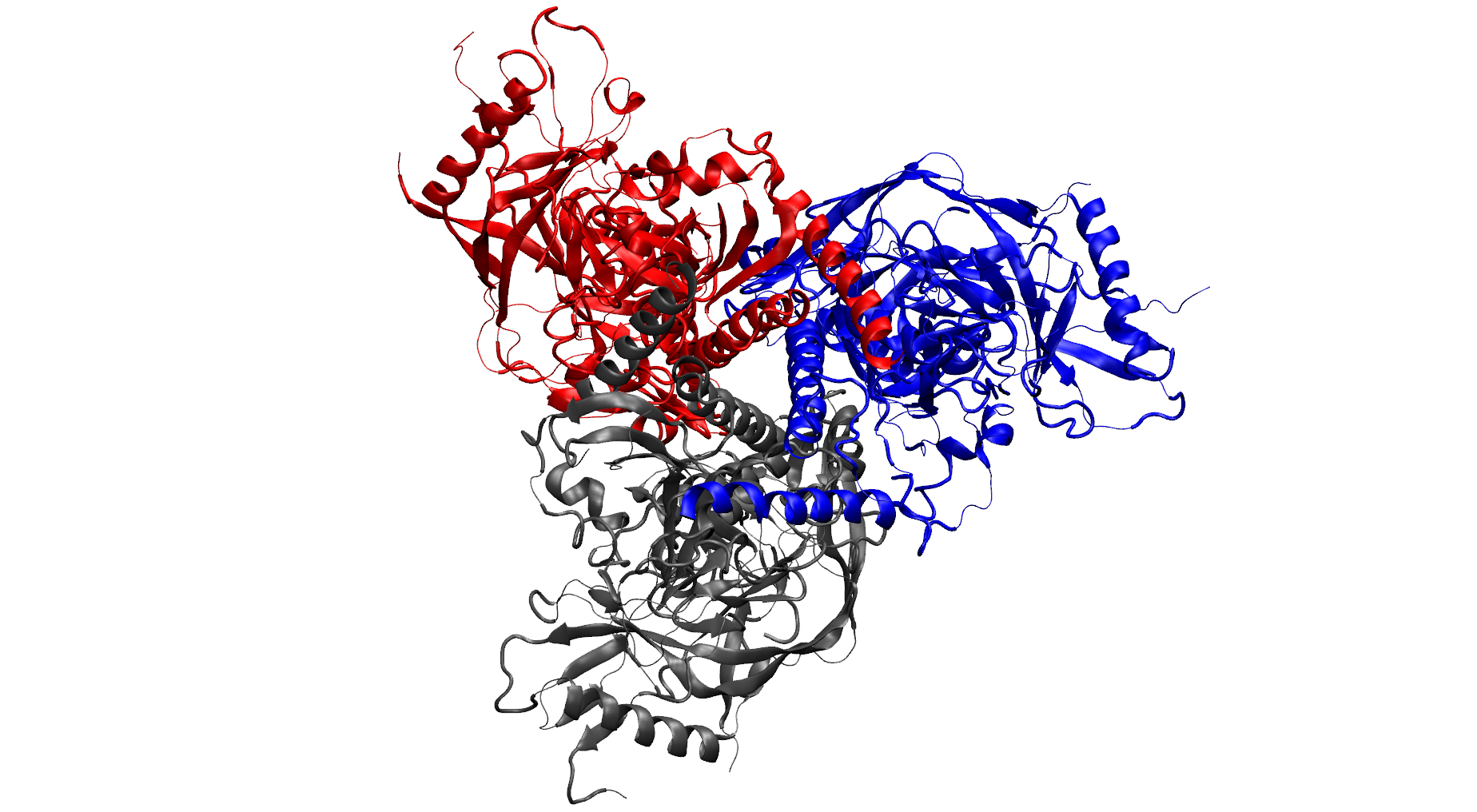
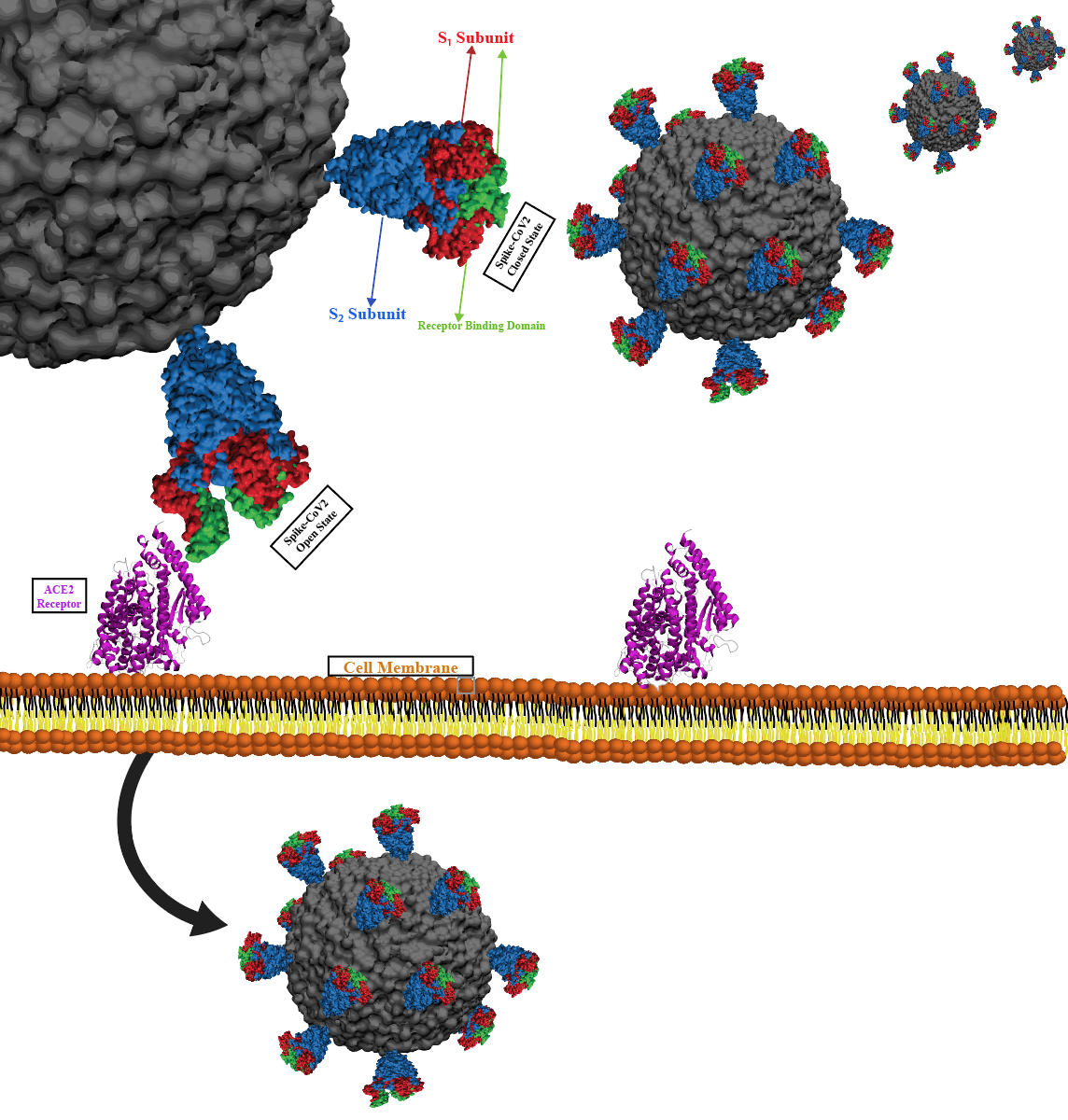
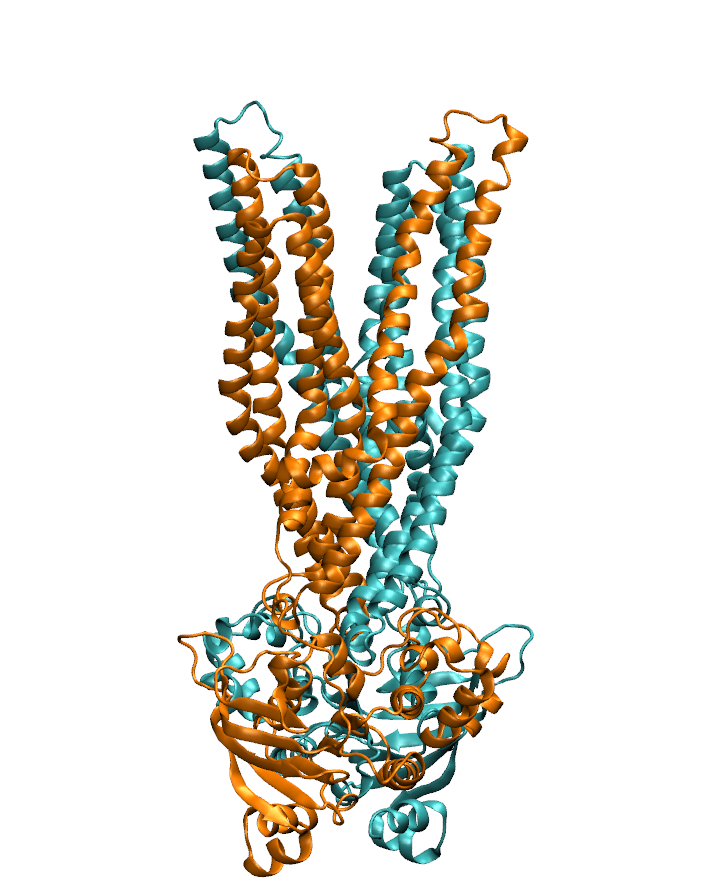
NSF I-Corps Award
Issued by the National Science Foundation, the
I-CORPS award was given in 2025-2026 for a Translation Potential of a Physics-based Computational Tool for Immunoassay Development (NSF IIP 2505779).
NIH Outstanding Investigator Award (R35), MIRA-ESI
Issued by the National Institutes of Health in 2022, Dr. Moradi has been awarded the
NIH Outstanding Investigator Award
for work on Physics-based characterization of functionally relevant protein conformational dynamics (R35GM147423) until 2027.
NSF I-Corps Award
Issued by the National Science Foundation, the
I-CORPS award was given in 2021-2022 for a Physics-Based Binding Affinity Estimator (NSF IIP 2138667).
NSF CAREER Award
Dr. Moradi has been awarded the
National Science Foundation CAREER Award in 2020 for work on Riemannian Reformulation of Collective Varaible Based Free Energy Calculation Methods
(NSF CHE 1945465) until 2025.
NIH R01 Grant
Granted by the National Institutes of Health in 2024, Dr. Moradi is a co-I for work on Probing Real-time Conformational Dynamics and Allosteric
Cooperativity of the HIV-1 Envelope Glycoprotein During Virus Entry in collaboration with Dr. Maolin Lu at The University of Texas at
Tyler until 2028 (NIH R01-AI181600-01).
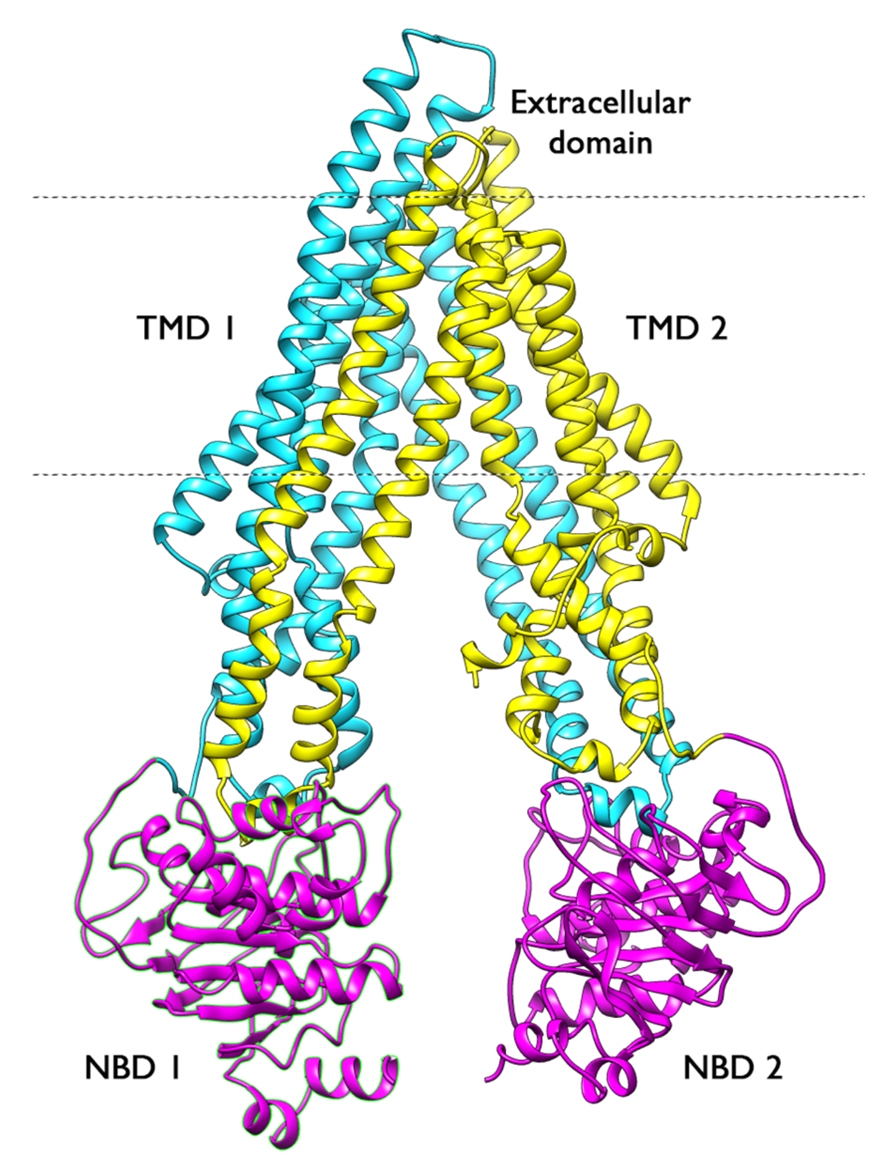
DOE Grant
Granted by the Department of Energy in 2020, Dr. Moradi was a co-PI for work on Protein Targeting to the Chloroplast Thylakoid Membrane: Structure
and Function of a Targeting Complex" in collaboration with Dr. Colin Heyes at the University of Arkansas until 2023 (DE-FG02-01ER15161).
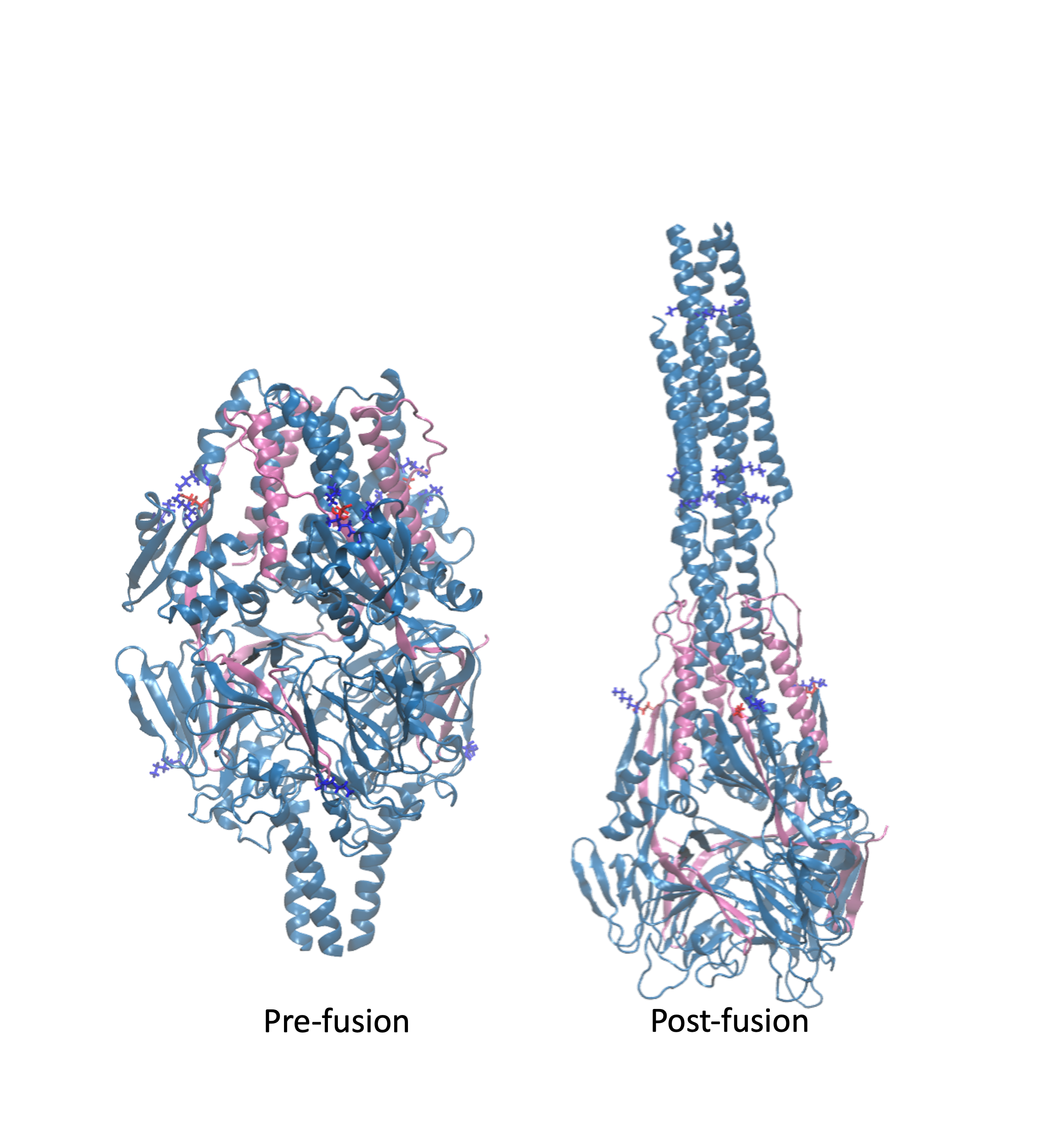
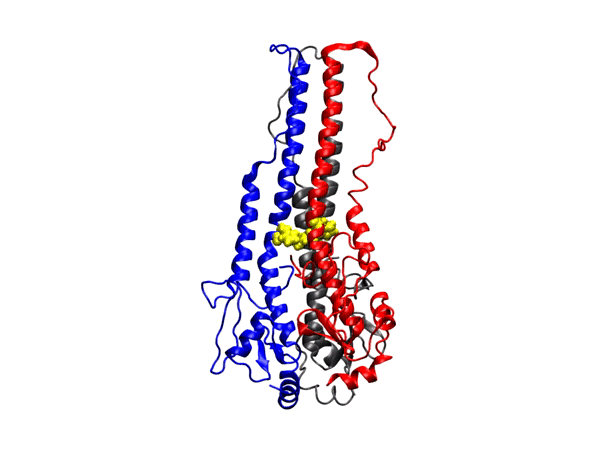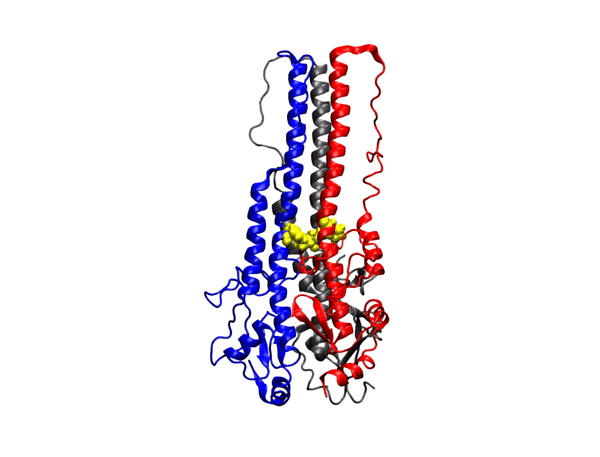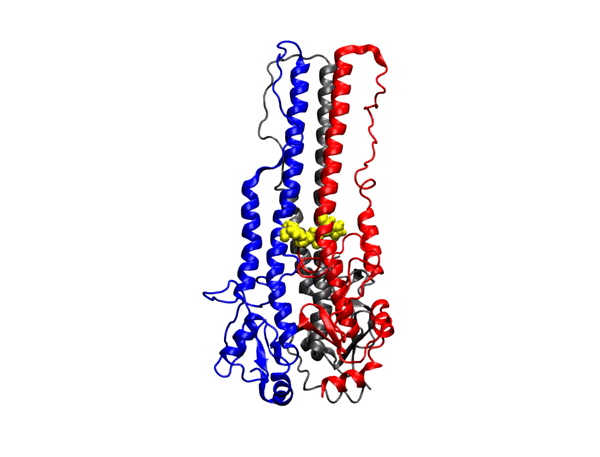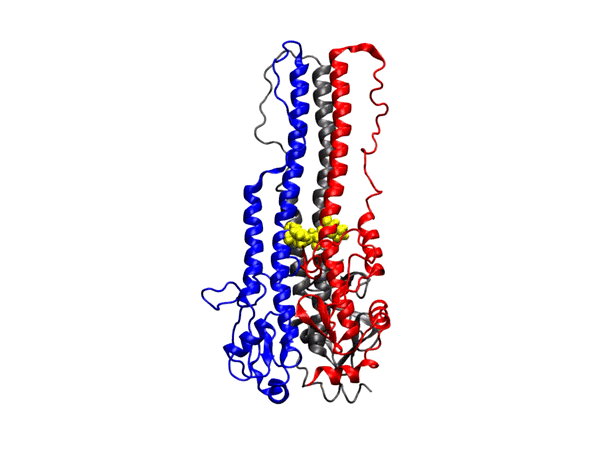
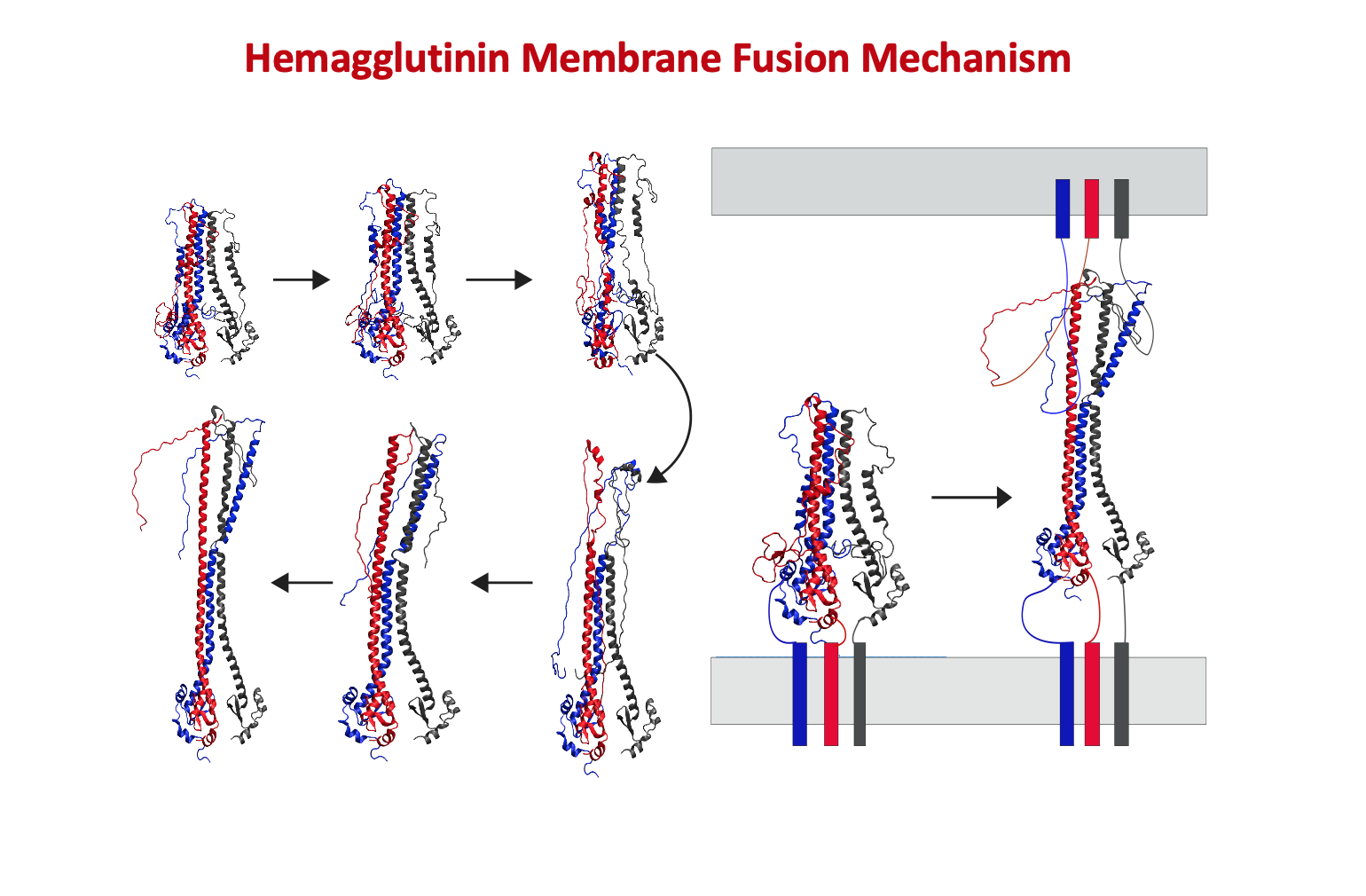
NIH R15 Grant
Granted by the National Institutes of Health in 2020, the lab recieved the NIH R15 Grant until 2023 for work on Molecular Characterization of the Influenza Hemagglutinin Mediated
Membrane Fusion (NIH R15-GM139140-01).
NSF HDR Grant
Granted by the National Science Foundation in 2019, the lab recieved the
NSF HDR Grant until 2022 for Collaborative Research: Atomic Level Structural Dynamics in Catalysts (NSF OAC 1940188).